Gliomas, among the deadliest forms of cancer, are brain tumors that present a significant challenge due to their rapid progression and resistance to treatment. Effective and early diagnosis is critical for improving patient prognosis. Deep learning, particularly through large-scale vision models like Segment Anything Model (SAM), offers a new pathway for tumor segmentation. This study seeks to address the primary challenge of adapting SAM for mp- MRI brain scans, which typically encompass multiple imaging modalities not fully utilized by standard three-channel vision models. We demonstrate that leveraging all available MRI modalities achieves superior performance compared to the standard mechanism of repeating a MRI scan to fit the input embedding. Our research also focuses on parameter-efficient tuning of SAM to effectively train the model while minimizing resource usage, showcasing significant improvements when evaluated across multiple datasets. Finally, we expose how SAM perceives differences across varied brain tumor domains by visually analyzing the features extracted on each of them. Our code and models are available at github.com/vpulab/med-sam-brain .

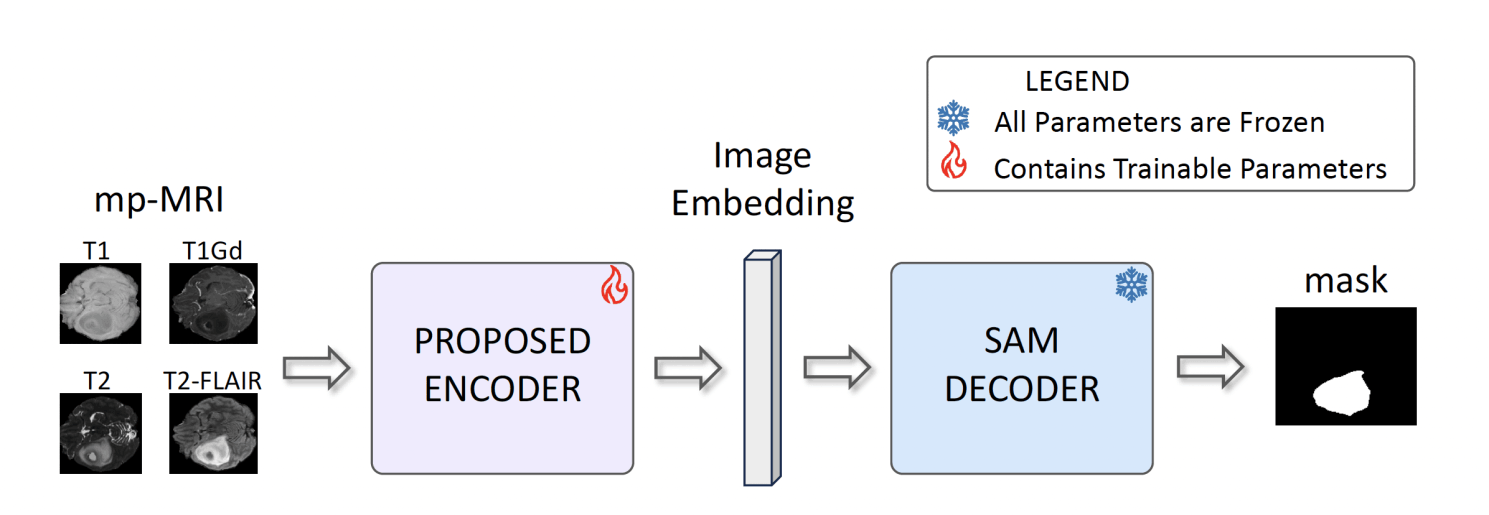
Pipeline overview. We propose to adapt the encoder by: 1) accounting for all the mp-MRI volumetric image modalities; and 2) specifically tuning of the encoder to retain the open-world segmentation capabilities of SAM.
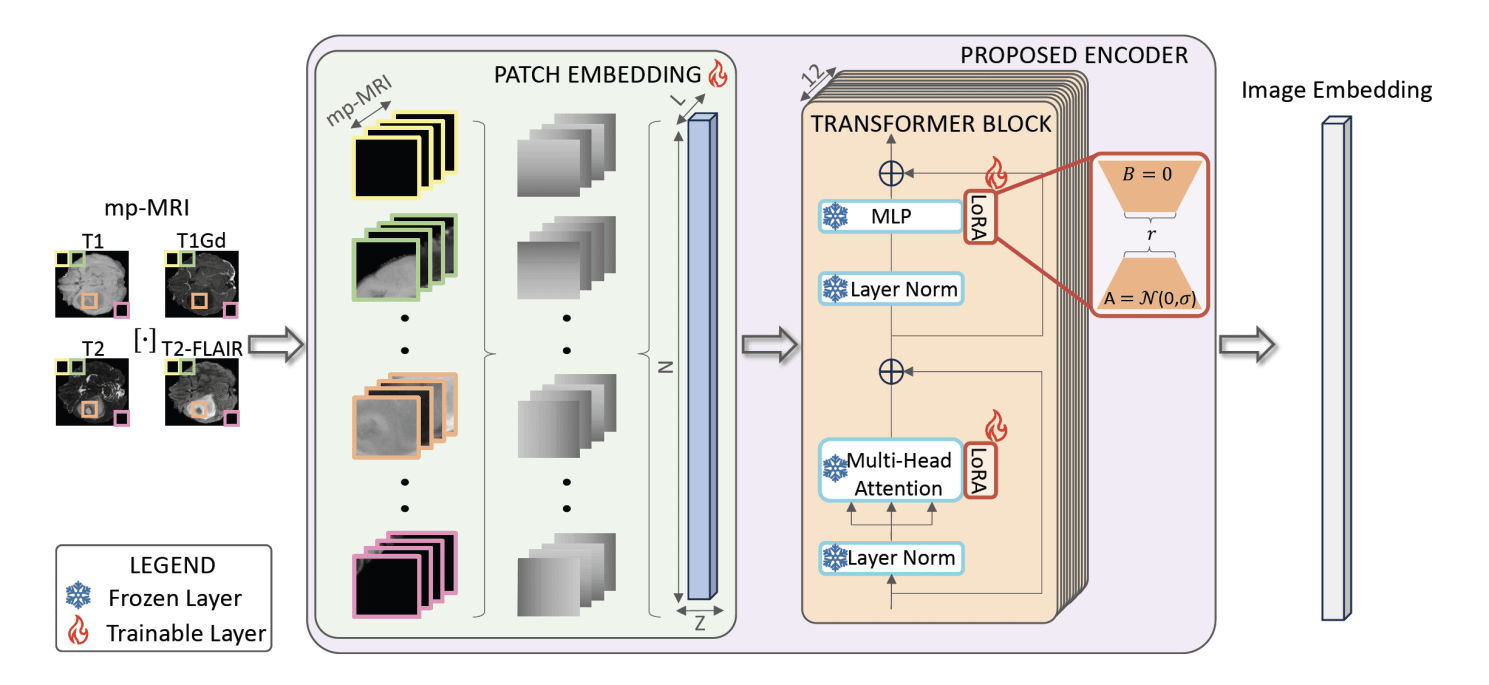
Proposed Encoder. We propose to modify the patch embedding layer, so that it accounts for the all the MRI modalities, allowing for a seamless integration of the information. Then, we employ LoRAs to tune Multi Layer Perceptron blocks (MLP) and Attention (Q,K,V embedding) layers of the transformer blocks.
Multi-modal adaptation.
Modified SAM’s encoder to process all four mp-MRI modalities (T1, T1Gd, T2, T2-FLAIR) jointly, including a cross-slice attention mechanism for 3D consistency.
Efficient domain tuning.
Applied parameter-efficient fine-tuning (LoRA) to adapt SAM to the medical domain while reducing computational cost and avoiding catastrophic forgetting.
Domain generalization analysis. Evaluated how the adapted model generalizes across different brain tumor domains (Adult, Pediatric, Meningioma, Sub-Saharan) and analyzed feature-space discrepancies.
Reference:
@inproceedings{diana2024sam,
title={How SAM Perceives Different mp-MRI Brain Tumor Domains?},
author={Diana-Albelda, Cecilia and Alcover-Couso, Roberto and Garc{\'\i}a-Mart{\'\i}n, {\'A}lvaro and Bescos, Jesus},
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition},
pages={4959--4970},
year={2024}
}
HTML Builder