Gliomas are aggressive brain tumors that require accurate imaging-based diagnosis, with segmentation playing a critical role in evaluating morphology and treatment decisions. Manual delineation of gliomas is time-consuming and prone to variability, motivating the use of deep learning to improve consistency and alleviate clinical workload. However, existing methods often fail to fully exploit the information available in multi-parametric MRI (mp-MRI), particularly inter-slice contextual features, and typically require considerable computational resources while lacking robustness across tumor type variations. We present GBT-SAM, a parameter-efficient deep learning framework that adapts the Segment Anything Model (SAM), a large-scale vision model, to volumetric mp-MRI data. GBT-SAM reduces input complexity by selecting fewer than 2.6% of slices per scan while incorporating all four MRI modalities, preserving essential tumor-related information with minimal cost. Furthermore, our model is trained by a two-step fine-tuning strategy that incorporates a depth-aware module to capture inter-slice correlations and lightweight adaptation layers, resulting in just 6.5M trainable parameters, which is the lowest among SAM-based approaches. GBT-SAM achieves a Dice Score of 93.54 on the BraTS Adult Glioma dataset and demonstrates robust performance on Meningioma, Pediatric Glioma, and Sub-Saharan Glioma datasets. These results highlight GBT-SAM’s potential as a computationally efficient and domain-robust framework for brain tumor segmentation using mp-MRI.

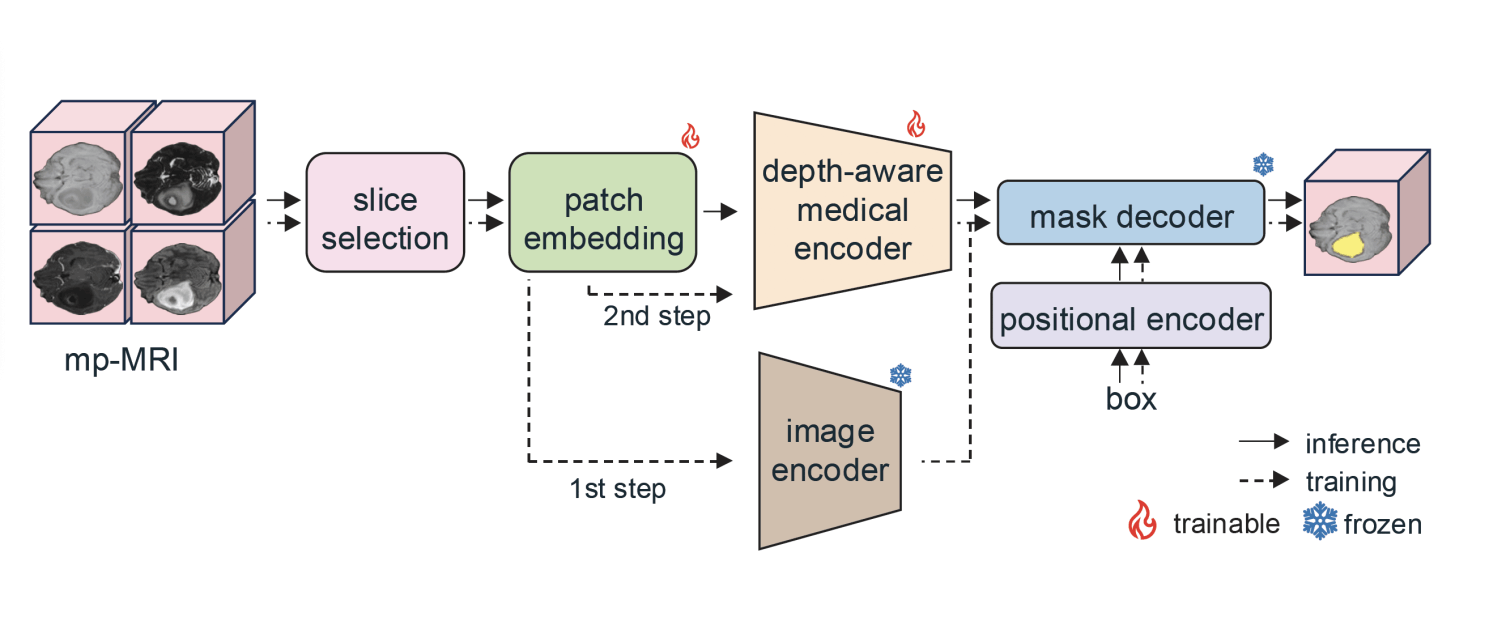
GBT-SAM pipeline. In a first training step, we perform slice selection to reduce computational costs while enhancing generalization capability. Moreover, the patch embedding layer is trained, while the rest of the modules remain frozen: image encoder, responsible for extracting features from the input slices; positional encoder, combining features with the bounding box information; and mask decoder, producing the predicted segmentation. In a second training step, the patch embedding layer is further trained alongside additional trainable components introduced in a modified version of the image encoder (depth-aware medical encoder): LoRA blocks and a Depth-Condition module.
Multi-modal Adaptation for mp-MRI: We adapt LVMs to process all four mp-MRI modalities (T1, T2, T1c, T2-FLAIR) jointly by modifying the patch embedding layer to accept 4-channel input and using a two-stage training protocol to capture each modality’s contribution.
Depth-Conditioned Correlation Modelling: To address the challenge of inter-slice correlation, we introduce a novel Depth-Condition block, which conditions feature representations on adjacent slices.
Parameter-Efficient Domain Adaptation: Recognizing that current models often lack domain-specific knowledge, we implement a parameter-efficient strategy to adapt the model to clinical imaging tasks.
- Currently Under Review in the journal Magnetic Resonance Imaging -
Reference:
@article{diana2025gbt,
title={GBT-SAM: Adapting a Foundational Deep Learning Model for Generalizable Brain Tumor Segmentation via Efficient Integration of Multi-Parametric MRI Data},
author={Diana-Albelda, Cecilia and Alcover-Couso, Roberto and Garc{\'\i}a-Mart{\'\i}n, {\'A}lvaro and Bescos, Jesus and Escudero-Vi{\~n}olo, Marcos},
journal={arXiv preprint arXiv:2503.04325},
year={2025}
}
Drag & Drop Website Builder